|
Image-based cytometry – a technology conquers cell culture
Image-based cytometry – a technology conquers cell cultureStar charts and Petri dishesCell cultivation has now become an indispensable part of scientific investigation and applications within medicine. From a simple substance test to the autologous cell therapies of the future, cell technologies are based on the controlled propagation of cells. An important step towards this goal is the non-invasive characterisation of the cell population grown. New methods in image-based cytometry offer an ideal platform technology for this purpose. Cells are everywhere. Nor do they serve us merely in our bodies – such as when reading these words, for example. They are also taking on an increasingly important role outside their host organism. Cell cultures are used in the laboratory, in medicine and in the cosmetics industry; they are even making inroads into such far-flung regions as sensors and the food industry. To function properly across this wide variety of applications, cultivation methods require constant improvement. One method that is as old as cell culture itself is that of describing cells by looking at them. The microscope is pretty much the oldest piece of equipment that is required for cell culture and yet – with its most recent models – still one of the most modern. One of the most exciting trends in cell culture is based on the “smart” use of microscopy and – more precisely – on the marriage of microscopy and computerised image analysis. Today, it’s a simple matter to create time series of cells in the Petri dish with the use of automated microscopes. And much can be learnt from these image sequences. Online monitoring of in vitro cell cultures The individual images in such series show an identical section of the Petri dish. A new generation of analytical algorithms now allows us to analyse the behaviour of the cells observed over the entire imaging time frame and at the level of single cells. Depending on the scientific issues being investigated and the experimental setup, several application scenarios are conceivable. One interesting application is the evaluation of chemotaxis assays, for example, which are used in drug research and drug testing. Another possible application is the real-time monitoring of proliferating cell populations. This online monitoring approach enables the visualisation of cell growth as a growth or confluence curve in real time. For quality assurance in the field of stem cell therapy, for example, it is imperative to perform quantitative observance, documentation and monitoring of behaviour and growth rates. To automate these processes, the Fraunhofer Research Institution for Marine Biotechnology (EMB) in Lübeck has developed a purpose-built software application capable of identifying each individual cell in the image data and calculating the cell count. Alongside the cell count, it is also possible to quantify the morphology of the cells identified, as well as determining cell size, length and shape. These morphological parameters can be presented as a function of time (cf. fig. 1). A cell population described using parameters that are sufficiently sound is termed a “parameterised cell culture” [1].
Fig. 1: Change to cell size depending on cell density. Cells in culture shrink to almost half their size when they become confluent.
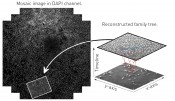
Go forth and multiply: migration and proliferation Yet image-based cytometry is capable of much more than describing cell cultures in terms of numbers: it also opens up a window onto these cells’ dynamic behaviour. Cells are not simply measured: their behaviour is also made capable of analysis. For this to be possible, cells must not only be identified in single images: each of them must also be followed through time. This procedure – termed “cell tracking” – is capable of measuring the cell’s pattern of migration, for example. The paths they take provide data on whether the cells move with purpose – such as with the objective of attacking a tumour, for example – or whether they instead just amble aimlessly around the Petri dish. Nor are the shapes of these cell trajectories the only things we can measure: we can also calculate the speed with which the cells move about. This data provides the starting-point for the kinds of subsequent analysis and modelling performed in chemotaxis assays, for example. Such assays not only enjoy broad-based use in oncological research, as mentioned above, but also in experiments addressing wound healing, which is definitively characterised by the migrational properties of the cells involved. Further scenarios include organogenesis, the investigation of infectious and autoimmune diseases, and research on regenerative processes. Nor is that all. Cell measurement precision can be improved even further by introducing the separate detection of cell divisions (mitoses). A “mitosis detector” of this kind has been constructed at Fraunhofer EMB. It detects cell divisions by the application of two distinct principles. First, a mitosis event is characterised by the morphological change occurring in the cell cycle just before division. The parent cell is drawn together by the spindle apparatus and assumes a ball-like shape that can be specifically detected [2]. The second possibility consists of searching for the characteristic optical changes present in a mitotic cell using a phase contrast microscope: the spherical cells are not only rounder but are also thicker than the flat adhered cells and so appear brighter in the phase contrast. With the aid of methods for machine-based learning, this mitotic pattern can easily be discovered in the image series [3]. Taken together, both forms of analysis – migration and proliferation – are already capable of delivering a very detailed picture of cell culture events. Complex processes can be investigated such as the metastasis of a tumour or the closure of a wound, for example. These processes are based on the intricate interplay of migration and proliferation, which are now capable of quantitative observation by a non-invasive, dye-free method. Haven’t we met before somewhere? We’re relatives, you say?! So far, we have been considering the behaviour of individual cells. Yet a Petri dish frequently contains millions of cells. Can we learn something about their “social behaviour”? Sure we can – by combining migration analysis (cell tracking) with the mitosis detector! The descendants of every cell undergoing division can be determined. This information can then be used to reconstruct the “family relationships” in the form of a cellular genealogical tree: this records all of the cell’s sisters, cousins and grandparents (cf. fig. 2). In addition, cells can also be identified that only “bumped into” each other during their lifetime, i.e. which had cell-to-cell contact – and possibly also exchanged information with one another.
Fig. 2: Family tree of a cell clone. The lower series shows three snapshots, over which the associated family tree is depicted, which classifies all of the cells in the clonal family.
By aggregating all of the morphology, migration and proliferation data described so far, we are then given the unique opportunity to compare cell behaviour in one cell family with the cell behaviour exhibited by other cell families. The reciprocal influence exerted by cell families on each other can also be investigated and visualised in detail. A star chart search: comparing cytometric data with protein expression patterns Although the methods described provide a wealth of data, they are still incapable of answering all of the questions posed by cell biology. One such issue is the question of the exact cell type involved, which cannot be determined from morphology or from migration/proliferation behaviour. Yet when investigating stem cells and questions of developmental biology, however, the resulting cell types are of paramount importance. As a rule, a molecular biologist obtains information about the cell type in question by detecting the presence of known functional proteins. Immunostaining is often used to make such detections. With this technique, a specific antibody binds to the functional protein; the bound antibody can be coupled with a dye that, in turn, renders the functional protein visible. It would clearly be ideal to combine immunostaining with image-based cytometry. To date, however, the integration of both techniques has faced a technical hurdle: while cytometric data can be recorded as part of the cell culture process, cells for immunocytochemical staining must be fixed – i.e. killed – before then being stained. In short, the two imaging techniques cannot be conducted at the same time: the staining process always gets in the way. How, then, we can find the cells observed with cytometry in the immunostaining images? Since these images are very different and contain a truly vast number of cells, it is not possible to simply pick out individual cells by eye or superimpose one image on top of the other (cf. fig. 3).
Fig. 3: The star chart problem. The cells whose family relationships were determined using cytometry (right) must be re-identified in the cell image produced by immunocytochemical staining (left). Help is provided by an algorithm adapted from the analysis of photos of stars in the night sky.
To overcome this obstacle, Fraunhofer EMB developed a new method with which identical cells in both types of image could be found and “matched up” by looking at their “cell constellations” [4]. The method resembles the technique developed in astronomy to permit the automated identification of stars and constellations in any random shot of the night sky [5]. To this end, constellations of three, four or five stars – or cells – are described using a unique hash value, which is calculated from the relative geometrical positions of the cells to one another. One condition for hash values is that they are stable, i.e. that each constellation is consistently described using the same hash value, independently of the rotation of the sky or Petri dish and the magnification chosen for the telescope or microscope. This is used as the basis for a search for the same patterns in both images – the final image in the cytometric image series and an image of the immunocytochemical staining. The basic structure of the search first requires the calculation of the hash values for all constellations of three, four or five cells in the immunostained image. With thousands of cells in the field of view, this might sound like a hugely demanding calculation, but it actually completes much faster than precomputation for astronomical constellations. Calculation of the hash values for the phase contrast image is then completed and the search begins for the most similar constellations in the immunostained image. Finally, the cells that make up these constellations are used to compute an image registry, with which the cell images can be congruently superimposed onto one another. The star chart search now makes it possible to integrate cytometric data such as degrees of cell relationship with any protein expression patterns chosen. This enables the investigation of truly fundamental questions within developmental biology whose answers previously required a much higher level of effort. In addition, it also permits the general automation of these methods and the execution of the observed processes as high-content analyses, and – in the future – an integration with systems biology. Outlook: Time-lapse microscopy as standard Enough, then, of what can already be achieved by using image-based cytometry. One minor drawback with the approach to date has been the fact that time-lapse microscopes are big, expensive and hard to integrate into routine cell culture. What’s stopping image-based cytometry becoming a runaway success in cell culture labs is a lack of inexpensive, rugged instruments that can fit into any incubation chamber and perform their tasks as unobtrusively as the chamber’s temperature or CO2 controllers. Fraunhofer EMB is already working on an “everyday” miniaturised cell scanner. The question is no longer whether but when image-based cytometry will become part of the standard toolbox used by modern cell culture laboratories.
Bibliography Picture: © istockphoto.com | cookelma |
L&M int. 1 / 2015
Free download here: download here The Authors:Read more articles online |