|
Scientist
>
Dr Marion Letzel
>
OMI (Organic Molecule Identification) in water using LC-MS(/MS): Steps from “unknown” to “identified”: a contribution to the discussion
OMI (Organic Molecule Identification) in water using LC-MS(/MS): Steps from “unknown” to “identified”: a contribution to the discussionIn a class of its own
The analysis of small organic molecules in water is highly demanding, Present-day analytical detection systems have not only shown rapid development but are also becoming more specific and more sensitive. One could of course imagine the task of testing water to detect molecules dissolved in it to be a very simple one – especially when using liquid chromatography (LC) coupled with (tandem)-mass spectrometry (MS(/MS)) [1, 2 fig. 1]. This conclusion is also suggested by the prevalence of LC-MS(/MS) equipment in water analysis labs. This coupling is indeed very successful when deployed for the quantification of known small organic molecules in water, as it is a superlative analysis method for molecules that are water-soluble and easily ionised. This is especially true if one can separate the molecules from one another chromatographically prior to detection with mass spectrometry [3, 4]. Such work involves the use of conventional reverse-phase liquid chromatography (RPLC; for medium-polar and non-polar molecules) and, more recently, hydrophilic interaction liquid chromatography (HILIC; for polar molecules) [5].
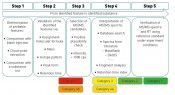
Fig. 1: Identification workflow, incl. categorisation of results
As a result of the increased use of accurate, high-resolution mass spectrometry (HRMS), qualitative analysis targeting the presence of organic trace substances is also possible. The task here is to analyse the sizeable dataset thereby generated, which presents analysts with substantial challenges: goal-oriented dataset analysis on the one hand and – in particular – the resultant analytical findings on the other. The original assumption – that mass spectrometry results are themselves sufficient to unambiguously characterise the molecules – proved ultimately untenable, not least due to the proliferation of isomeric compounds [6]. Current consensus states that, without further background knowledge, complex samples of this type are effectively beyond comprehension or evaluation [2, Fig. 2]. Unavoidably, therefore, one must evaluate the results obtained before then assigning them into categories. Categorisation takes place depending on the level of organic molecule identification (OMI) and the strategic orientation. Accordingly, the category thus results from the analytical method used on the one hand but also from the databases and reference standards used on the other.
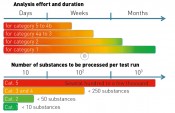
Fig. 2: Analysis effort and duration
Baseline situation for OMI categorisation In recent years, the analytical techniques and strategic approaches within water analysis – while initially discrete – have moved slowly but surely to a more uniform perspective. In the water community, there is consensus not only on the identification strategy with instrumental analysis but also on the use of databases and reference substances for the full identification of trace substances. Just recently, the team headed by J. Hollender published a classification of measurement results that is subdivided into levels 1 to 5 (incl. 2a/2b) [7]. The assignment is primarily based on the use of high-resolution, accurate-mass mass spectrometry. Other details are also considered as part of the evaluation strategy. Readers should take the time to consult this “Viewpoint” article since knowledge of its content is assumed by discussions in the sections below. The complementary categorisation presented here accords with the one referred to above. Supplementing this approach with comparison testing from multiple (here: two) labs extends it while also employing it as a further basis for interpretation. Principles of OMI categorisation with multiple associated laboratories Organic molecules are viewed as being identified in the proper sense of the word when one has decoded these initially unknown substances to the extent that one was able to produce a reference substance and confirm this physically/chemically by using synthesised reference materials. Following the definition of this OMI as category 1, category 2 is then utilised for two types of molecule without an explicit reference substance: ones unambiguously assignable either via analytical chemical data or with the aid of substance databases (2a); or ones that can be viewed as confirmed via what are termed “diagnostic fragments” from the mass spectrometry used (2b). If both criteria are satisfied, then classification of the OMI as category 2 can be maintained until the OMI is promoted to Category 1 by comparison with a reference substance. While category 2 can be achieved by a single laboratory (or mass spectrometry system), it can also be obtained as a result of combining the data from multiple laboratories (or multiple mass spectrometry systems). The OMI is assigned to category 3 in cases where even the participation of multiple labs (or mass spectrometry systems) does not generate consensus on an unambiguous molecular structure. Such cases occupy a “grey area” within identification, so to speak: while some evidence speaks for the OMI concerned, this evidence is not entirely unambiguous. While the participation of multiple laboratories can corroborate a conjecture, it may not necessarily produce unambiguous evidence. In this process, an MS/MS result can be coupled with the accurate mass from a second mass spectrometer. If the second system does not refute the structure – e.g. via a different molecular formula – it can then be assigned to category 3. Category 4 is subdivided into two sub-categories (4a and 4b). By definition, a substance is typically assigned to category 4 (a or b) in cases where a) it could be measured only by a single mass spectrometer (incl. MS/MS measurement) or b) could be determined by multiple laboratories (accurate mass, but without any MS/MS results). As such, the use of suitable additional laboratories (or technologies) can mean category 4 is rapidly superseded, with promotion possible to at least category 3. To category 5 are assigned what are termed “masses of interest”. This category can also contain (non-) accurate masses of molecules from individual laboratories. Categorisation using the following levels of organic molecule identification (OMI): // Category 1: OMI confirmed by reference substance
// Category 2: OMI via multiple unambiguous indicators (without reference substance), possibly with restrictive subdivision: 2a: OMI unambiguously confirmed solely by using databases // Category 3: OMI with results from multiple laboratories (incl. MS/MS and accurate mass) but without unambiguous indicators
// Category 4: OMI with initially non-assignable results from 4a: from a single laboratory (incl. MS/MS) // Category 5: OMI with “masses of interest”, which are non-assignable
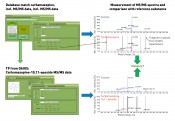
Fig. 3: Database search and subsequent identification of database matches using LC-MS/MS
Strategic methodology for OMI categorisation
What, then, do we need for the categorisation of analytical data? First, we establish an analysis on the basis of LC-MS/MS. Identification strategies are also available for other separation/detection techniques, with correspondingly similar variants. The strategy described in detail here defines a workflow comprising five steps. The category to which a “mass of interest” can be promoted in the course of these five steps depends on the results obtained within each individual work step. Note: the five categories used for an identification do not correlate with the five work steps to be performed (see fig. 1). Step 1 comprises the fundamental data processing, such as “feature” recognition, accounting for blank values and graphical reproduction. In this step, blank value correction is to be seen as the most important step, taken in order to filter out false positives. The features determined (exact mass peak with corresponding retention time) can then be compared even during this step with a database, in order to obtain some initial suspected compounds. If this option is skipped, then the features are processed further without assignment to suspected compounds. Step 2 involves plausibility checking of the analytical data obtained in step 1, with the aid of this data's physical and chemical properties. Features without the additional information of a suspected compound can be assigned to a molecular formula here, by taking into account the exact mass and the isotope pattern. In step 3, predetermined parameters are used to select features that are referenced for the separate performance of experiments using (tandem)-mass spectrometry (e.g. QTOF-MS). Here, the selection of the threshold (i.e. the minimum height of the selected peak) is a key parameter, necessary in order to obtain meaningful and sufficiently high-intensity production spectra. This step is also the first in which the OMI categorisation is decided, since the MS/MS spectrum constitutes an essential decision-making aid for assignment. If recording of MS/MS data is possible in only one participating lab, then the only possible assignment is to category 4b. Category 5 contains all components for which no fragment ion spectra could be obtained. At this stage, it is also entirely possible to record qualitative data from regular samples by using a triple quadropole mass spectrometer. Assuming the MS units stem from the same maker and have identical source geometry, this involves applying the fragment masses recorded in the product ion spectrum for identification to a triple quadropole method with corresponding LC method. This offers a highly-sensitive and selective procedure for the qualitative determination of unknown substances even before final assignment to a category with modest effort for larger numbers of regular samples. At the same time, this enables an initial prioritisation by environmental relevance. Currently, step 4 is still time-consuming and may also require additional measurements to obtain meaningful results following interpretation of the measurement data. The results from this step are definitive for the identification of a compound, since they form the basis for decision-making as to the reference substance with which step 5 must be performed. Further validation typically utilises data from separate laboratories; this requires additional effort in terms of time and coordination. If step 4 is successful, then the 5th step can proceed with the unambiguous assignment of the previously unknown substance. This requires the availability of an existing or – in the more complex case – newly-synthesised reference substance. Assignment of OMI categorisation The time taken depends on the category and is highly variable (see fig. 2 Top), ranging from just a few days to as long as several months for the full and complete identification (category 1) of a compound. The procurement or synthesis of a reference standard is one of the most time-consuming and costly steps in this strategy. At the same time, the number of components to be processed falls drastically during the course of analysing a sample, from the first step (feature finding) through to potential identification by use of a reference substance (see fig. 2 Bottom). The scope of component reduction work is strongly dependent on the choice of parameters during plausibility checking (step 2) and the selection of candidates for additional experiments, such as the recording of product ion spectra. The extent to which a dataset must/can be reduced is decided in accordance with the number and intensity of components obtained by feature finding. It is certainly advisable to select the highest-intensity or most striking components for further analysis. A useful aid to initial decision-making here is to query the matches in substance lists and to use the associated metadata, such as (e.g.) toxicity data, degradation rates when evaluating technical processes or specific issues of interest arising from the context of the investigations performed. One example of such a process-driven issue might well be the discovery of theoretically determined transformation or degradation products for previously identified compounds during a specified process. Application of OMI categorisation
– using the example of carbamazepine and its transformation product The methodology for database searching and further analysis of regular samples is based on an LC-QTOF screening measurement with positive electrospray ionisation. Suitable application software is used to extract features from the MS scan data. These features are characterised by their exact mass and retention time. A comparison of the features detected with a materials database such as STOFF-IDENT [8] or DAOIS [9] using the exact mass results – in our sample case – in a match for carbamazepine. By consulting the database entry for carbamazepine in DAOIS, the corresponding transformation product (TP) carbamazepine-10,11-epoxide entered is then used for investigating the raw data. Prediction tools such as the University of Minnesota's Predicted Pathway System (UM-PPS) [10] also offer a further option for locating transformation products. These proposed formulas and the exact masses that are calculated from them are used for the investigation of the raw data. In this way, further analysis not only uses suggested known starting compounds – sourced from the database searches – but supplements this on an ad hoc basis with potential transformation products. Identification of the starting compounds and their TPs is now conducted jointly using the same analysis strategy. Once the defined parameters have been satisfied in terms of plausibility checking and intensity, the next step is then to record and interpret the product ion spectra. The (tandem)-mass spectrometry results then serve as the basis for assigning the substances to the respective categories. By recording the MS/MS spectra solely in the regular sample and without measurement of the MS/MS spectra for a reference substance, the fragment masses listed in DAIOS would already enable an assignment of both substances to category 2. The highest degree of confirmation (category 1 – see figure 3) is achieved if the reference substance is present, and product ion spectra are recorded for both sample and standard – thus enabling confirmation of the compound. Summary An OMI categorisation based on LC-MS(/MS)-based data obtained from water samples is possible from a number of perspectives. One such perspective published recently takes as a starting-point a lab and its facilities, i.e. is oriented on the mass spectrometer, databases and/or reference substances. The categorisation presented in this article also takes into account the analytical environment of individual laboratories, utilising the association of this – in the “best case” – supplementary and complementary information. To be able to compare data, one crucial factor in this approach is naturally the adoption of shared standards by the laboratories. Retention times on dissimilar columns can thus be harmonized or – even more usefully – TOF-MS data (besides the accurate mass) can be validated by QTOF data and extended by tandem MS measurements (with structural information) as required. Experience has shown that the relaying of these MS/MS data is a useful tool in the qualitative analysis of unknown substances in regular samples with the aid of a triple quadrupole method. Whether case-based or generic, both databases and reference standards can be both shared and updated very simply among networked laboratories. This is another reason why we firmly believe that the future of screening techniques is to be found in the networked lab environment. Ultimately, this will lead to the adoption of lower-end LC-MS systems with low-resolution MS technology by routine lab testing, with these units being extended by expensive, high-resolution, MS systems as needed. In addition, work on producing a shared data repository in this field will have the effect of increasing the informational density of existing substance databases, thus further improving the professional use of such databases. We are looking forward! Bibliography
[1] Türk, J. (2012) Labor&More, 8, 46–49 Picture: © istockphoto.com | Jag_cz |
L&M int. 4 / 2014
Free download here: download here The Authors:Read more articles online |